|
Lesson Nineteen
by Dr Jamie Love |
|
 and
and  licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.

|
Lesson Nineteen
by Dr Jamie Love |

|
By now you are probably just dieing to learn all the nuts and
bolts of genetics - the "molecular stuff"! That's what
this section is all about. We are now entering the fascinating
world of "molecular genetics". In this and the next
five lessons you will learn about genetics as perceived at the
molecular level. That is not to say that everything else you have
learned is no longer relevant. We will still be referring to mitosis,
phenotypes, recessives and other important concepts.
Molecular genetics is an extremely important and relevant subject.
I think you will agree that I have saved the best for last. 
In this lesson I will teach you all about the molecule that carries, from generation to generation, all the information to make an organism. That molecule is called "DNA" - an abbreviation for "deoxyribonucleic acids". You will learn all the details about DNA shortly but first I want to tell you (a little story of) how "standard" genetics evolved into molecular genetics. This history is important because it acts as a bridge between the genetics you have learned and the new kind of genetics we are now entering.
The Idenitification of the Genomic Molecules
Stucture of Nucleic Acids
Stucture of Nucleic Acid Polymers
In 1908 Sir Archibald Garrod, a British physician, suggested that some human diseases were caused by "inborn errors of metabolism". He explained that the improper functioning, or the complete lack, of a specific enzyme caused specific diseases and many of these diseases were inherited. Parents who had the genetic information producing a bad enzyme, or none at all, passed that defective gene to their offspring - who would express the same (diseased) phenotype. Garrod believed that something was "defective" with the genetic information needed to make the normal enzyme.
Leap ahead a few decades to 1941 when two biologists, Beadle and
Tatum, were studying a common fungus called Neurospora.
Normal Neurospora can grow well in a standard "growth
medium" (a broth) even when that medium lacks vitamin B6.
Beadle and Tatum irradiated Neurospora with X-rays and produced
"mutants". Mutants are defective in some genetic way and these particular
Neurospora mutants died unless their medium was supplemented with B6. The experiments
allowed Beadle and Tatum to produce (by X-rays) and identify (by "selective
growth medium") mutant Neurospora cells. They concluded
that the X-rays had changed, mutated, a gene in each of these individuals that encoded an
enzyme for vitamin B6 synthesis.
That is just a simple conclusion from a simple experiment and it would not mean a great
deal to you or anyone in genetics. However, Beadle and Tatum took
their simple observations and explanation further by suggesting
that a particular gene codes for the synthesis of one particular
enzyme. This became known as the "one gene - one enzyme hypothesis"
and it is the foundation of molecular genetics.
Subsequent genetic
and biochemical experiments by others supported the hypothesis
and extended it beyond enzymes to include all proteins so it was
soon renamed the "one gene - one protein hypothesis".
This hypothesis was supported by more experiments and was unchanged
for a couple decades. Then scientists discovered that many proteins
are made of two or more separate polypeptides (polypeptides are
short sections of protein) that are often coded by different genes.
These different genes could be, and often are, scattered throughout
the genome but their products (polypeptides) come together to make
the final functional protein. That means the situation is not
as simple as Beadle and Tatum had thought. A single protein, produced
by different polypeptides coded by different genes at different
loci, means that the final affect, the phenotype, could be under a complicated
control system that resembles epistasis and multigenes! So a better
way to think about the Beadle and Tatum model is to think in terms
of "one gene - one polypeptide". However, even this
is not a truly complete picture of how a gene's function should
be defined at the molecular level. We now know that not all
genes code for proteins or peptides. Genes code for
RNA (an abbreviation for "ribonucleic acids", which
we will discuss shortly) and some RNAs, all on their own, can
produce phenotypes just as well as an enzyme, protein or peptide!
Therefore, the current model is best described as "one gene
- one product". (Phew!  )
)
The model proposed by Beadle and
Tatum "one gene - one enzyme", extended by others to
its modern form of "one gene - one product", defines
the critical link between classical genetics, that you studied
in our first three courses and molecular genetics which you will
study now.
OK, that is how classical genetics evolved into molecular genetics.
DNA is the "magic molecule" found at the intersection
of these two disciplines. The molecule called deoxyribonucleic
acid (DNA) was first described in 1869 by a man named Miescher
but his was a simple chemical description that did not seem to
have any relevance to genetics. Indeed, until the middle of the
20th century, most biologists and chemists believed that proteins
were the genetic code! It sort of made sense - even the revised
versions of the model proposed by Beadle and Tatum focused on
proteins and the huge diversity of proteins seemed natural to explain the complexity of genetics.
However, a series of experiments conducted with bacteria finally
proved that nucleic acids, not proteins, are responsible for genetics.
|
The bacterium Streptococcus pneumonia has a variety of
strains - some are harmless and others lethal - and it was clear
from previous work that these phenotypes are under genetic control because the phenotype, harmless or lethal, is inherited.
In the late 1920's Frederick
Griffith mixed dead (heat-killed) lethal bacteria with live harmless
bacteria and injected the mixture into mice. The mice died!
Griffith concluded that there was a "transforming material" passing from the dead lethal bacteria to the live harmless bacteria, "transforming" them into lethal bacteria. These "transformed" bacteria remained lethal and passed the lethal trait to their offspring, therefore, this "transforming material" is the genetic material. But, Griffith believed (as did everyone else) that it was a protein being passed in the transformation. | 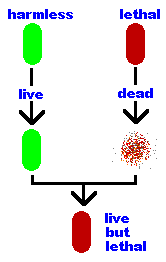 |
|
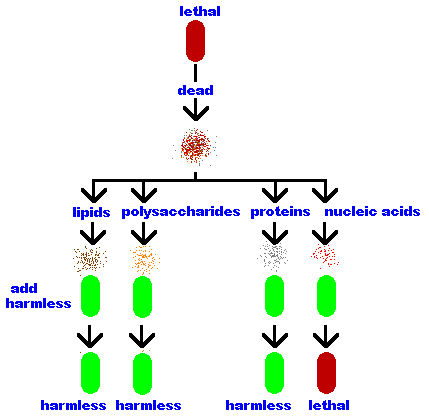
Oswald Avery and his co-workers built upon Griffith's work.
He lysed (broke open) the lethal bacteria and separated the bacterial
components into its parts - lipids, polysaccharides, proteins
and nucleic acids. Then he repeated Griffith's work but this time
trying each component separately. He discovered that only
the nucleic acids transformed the harmless bacteria into
lethal ones.
That was not the end of Avery's work. He knew, as you will soon
learn, that there are two types of nucleic acids - DNA and RNA. Avery
repeated his experiment but this time he treated the nucleic acids
with an enzyme that destroys RNA while leaving the DNA unaffected.
This "RNA-free" solution of nucleic acids was still
able to transform harmless bacteria into lethal bacteria so Avery
proved that DNA is the genetic material!
|  |
So, what is DNA?
James Watson and Francis Crick discovered the structure of DNA, using important data collected by Rosalind Parkers, and published their discovery in 1953. The details of that discovery make for a wonderful story and you can read Watson's personal account in his (excellent, short and understandable) book, "The Double Helix". I will not tell you that story.
Instead, I will leave history behind and jump right into a detailed description of the molecules of genetics! A detailed understanding of these molecules and the structure of DNA is required in order to understand the most important ideas and technology of molecular genetics
All genetic information is contained in nucleic acids and these are made of three types of molecules …
|
Carbon 1 (C1) is where the base is attached. More on that later.
Carbon 2 (C2) tells you if it is a ribose or deoxyribose. In deoxyribose "de oxy" is missing. 
Carbon 3 (C3) is the point of attachment for more nucleotides through an oxygen (actually the hydroxyl, OH). More on that later, too. Carbon 4 (C4) completes the ring via an oxygen (O) which bridges to the carbon 1 (C1). Carbon 5 (C5) hangs away from the ring and is the point of attachment for its phosphate(s). More on that later, too. | 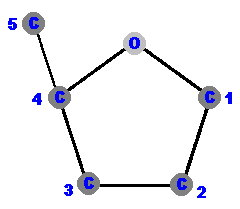 |
|
This molecule is a ribose because it has an oxygen, actually a hydroxyl (OH), on C2. (Right?)
It bears repeating, because some students miss this point and get confused, that the oxygen between Carbon 4 and Carbon 1 completes the pentagon shape BUT these are called "pentose sugars" because they have five carbons, not five "sides". (Don't worry about the hydrogens near some of the carbons. They are just there to complete the molecule but most of them are not important.) |  |
|
I want you to imagine this sugar as a cartoon person with the
Carbon 5 as his right hand extending upwards and away from the "body"
of this pentagon-shape person.
He has a big "phosphate mitten" on his right hand. Imagine the oxygen as his head and the C1 as his left shoulder. It is on his left shoulder that he will have attached the base - which acts as his other hand. The hydroxyls (OHs) dangling down from C2 and C3 are his feet! A "deoxy-person" would be missing his left foot (but his C2 "hip" is still there so even a deoxy-person has a complete sugar body)! This cartoon might seem like a silly way to represent a sugar molecule but, as you will see, it is a useful learning tool and handy reference chart for this important molecule. The phosphate is a standard PO4 group - a single phosphorous atom surrounded by four oxygen atoms. Free-floating phosphate has a charge of negative 2 (-2) and is represented as PO4-2. However, phosphate in DNA is not free phosphate. It is (covalently) attached to the C5 of the sugar via his right arm, and that reduces its charge to negative 1 (-1) so it is represented as PO4-1. | 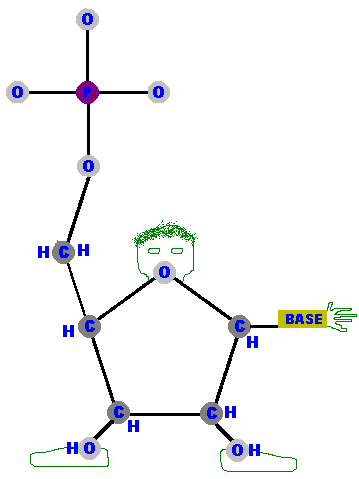 |
(Please note that these descriptions are only meant to help orient
you in this complex molecule. More advanced students might worry
about the exact accounting of the oxygens, hydrogens and charges but to
explain the accounting would require you to learn more details
of chemical reactions than are necessary for this course. So,
don't worry about it.  )
)
Phosphates and sugars make up the "backbone" of DNA and RNA. These two molecules can (and do) form a chain that runs from a phosphate on the previous molecule, to C3 (the right foot and hip) through C4 (the right shoulder) to C5 (the right hand holding a mitten for grabbing the next molecule). The chain continues with the C5 phosphate (which is attached to the C3 of another sugar - and so on). We'll come back to that soon but before carrying on, have another look at our cartoon person and run your finger along the backbone I have just described.
Now let's tackle the ever-important bases. There are two types of nitrogenous bases.
are a single hexagonal ring of carbons and nitrogens. 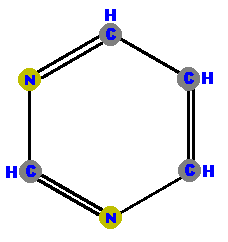 Pyrimidine | are double rings made of a pyrimidine with a pentagon added.  Purine |
The carbons and nitrogens in these molecules are also numbered but it not important to learn each of them. Instead, simply accept that we use a numbering system to identify each part in the rings that makes up these two nitrogenous bases.
At this point you may be wondering, "If the pentose sugars are numbered one through five, does the numbering of the carbons in the bases start at six?" The answer is "no", we start numbering from one again. Of course, that could cause some confusion. ("Are you talking about C1 in the sugar or C1 in the base?") Therefore, chemists decided to call all the carbons in the sugar "prime" (meaning first) atoms. So we refer to the carbon 1 in sugar as "carbon one prime" and the carbon in the bases simply as "carbon one". Indeed, most chemist no longer even mention the carbon and would just say "one prime" when speaking about the C1 in the sugar. That means the C3 of the sugar's carbon (the right hip) is called "3 prime" and the C5 of the sugar's carbon (the right hand) is called "5 prime". From the bottom of his right foot to the top of his right hand, our cartoon man runs from 3 prime to 5 prime. To simplify the writing, we use the abbreviation ' (a little dash or an apostrophe) to signify prime so the sugar runs 3' to 5' with the 4' shoulder in between.
DNA has four different bases (and abbreviations).
Adenine (A) and guanine (G) are purines which are distinguished and identified by the oxygen (O) or ammonia (NH2) attached at specific positions. Make a note where these "side groups" are positioned so you can identify these two molecules, and distinguish between them. (The astute student may also note how double bonds have shifted and affected the resonance patterns of the guanine molecule, but that requires a higher level of chemistry to appreciate - so don't worry about the bonds.)
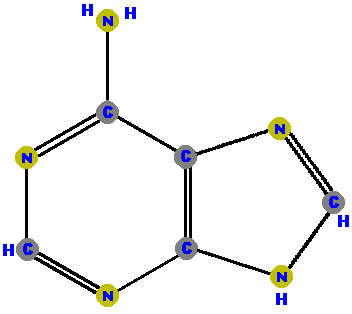 | 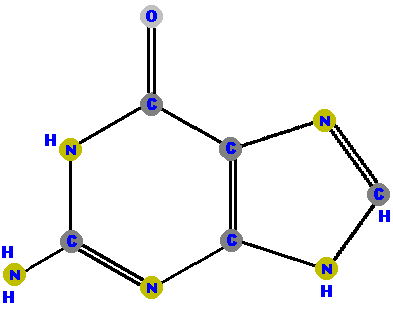 |
Thymine (T) and cytosine (C) are pyrimidines. They are distinguished and identified by the oxygen (O), ammonia (NH2) or methyl (CH3) attached at specific positions. Again, make a note where these "side groups" are positioned so you can identify the molecules later.
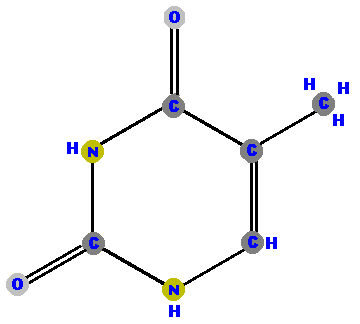 |
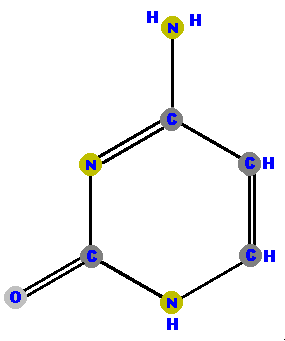 |
|
RNA (ribonucleic acid) does not have thymine. Instead, it has
the base uracil (U).
Notice that uracil is thymine with the methyl (CH3) removed. | 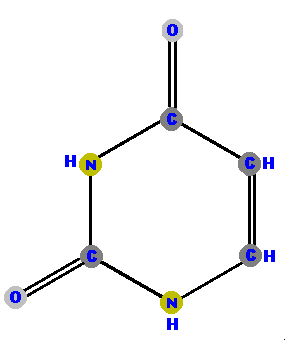 |  |
You now know the tiniest details to understand the larger molecules so let's put these smaller molecules together to make nucleic acids and along the way learn some more structures, definitions and vocabulary.
Sugar's carbon one (1') is the site where bases are attached.
A specific atom in the ring's base is used as the point of attachment
but we won't need to know the details of that positioning.
Any base (A, G, C, T or U) can attach to either sugar (ribose
or deoxyribose) to form a "double molecule" called a
nucleoside. The bond linking these molecules (sugar and
base) is called a glycoside bond.
If the sugar is ribose we have a ribonucleoside but if
the sugar is deoxyribose we have a deoxyribonucleoside.
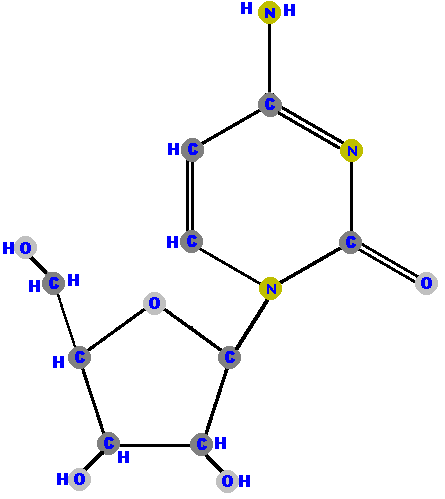
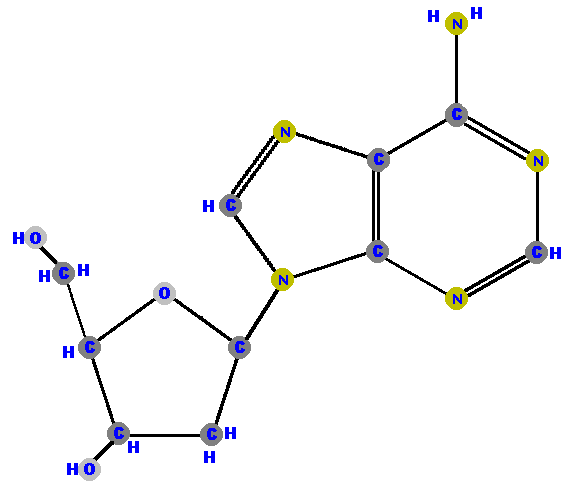
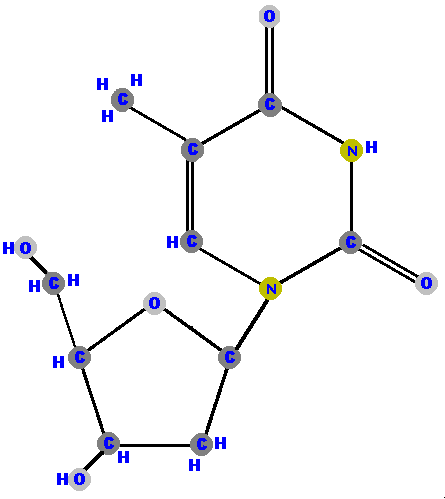
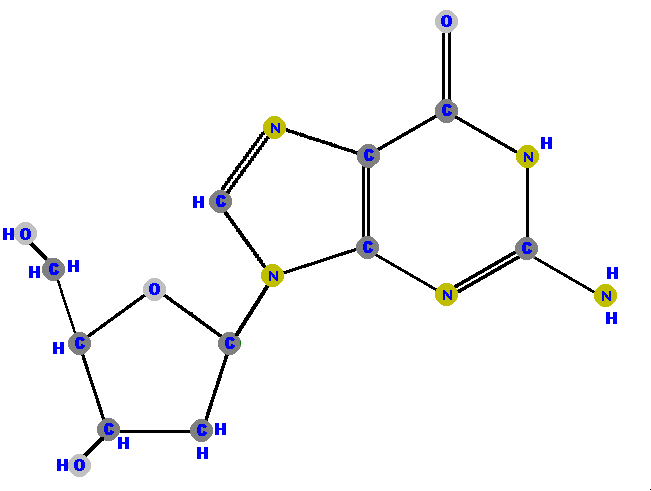
If the attached base is adenine the molecule is called "adenosine" or "deoxyadenosine" depending upon which sugar is attached (ribose or deoxyribose, respectively). When guanine has the ribose attached it becomes "guanosine" and with the deoxyribose attached it is called "deoxyguanosine". Cytosine becomes "cytidine" or "deoxycytidine" when the appropriate sugar is attached. Uracil is only found bound to ribose and that molecule is called "uridine". On the other hand, thymine only binds to deoxyribose and it is then called "deoxythymidine".
Don't worry about memorizing all these names (yet  ). Just be aware that
they take on different endings. I will list them all shortly in
a summary table.
). Just be aware that
they take on different endings. I will list them all shortly in
a summary table.
Here (below) are the four ribonucleosides. Notice that they all have a
hydroxyl group (OH) at the C2' position. Also, notice that thymine
is not represented among the ribonucleosides because uracil "takes
its place" (as you will see shortly).
[These images can get to be very large and it becomes inconvenient for you to spend a lot of time scrolling up and down, left and right, to see the whole molecule (if you have a standard size computer screen). Therefore, I will use a "hypertextbook trick" to scale the images down on your screen but you can see the details of each molecule by simply clicking on it. After you have studied those details, use your browser's "back" button to return to this screen.]
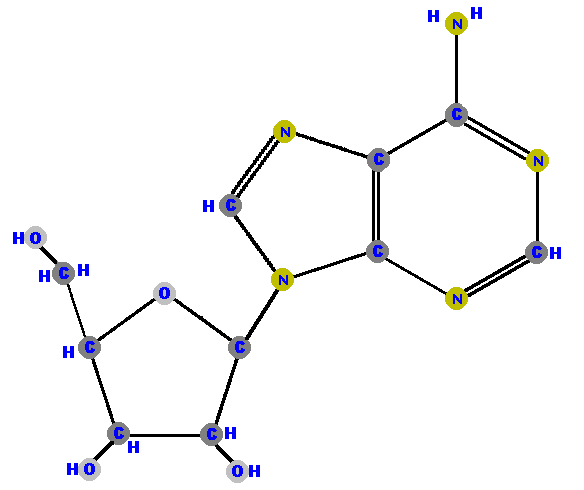 |
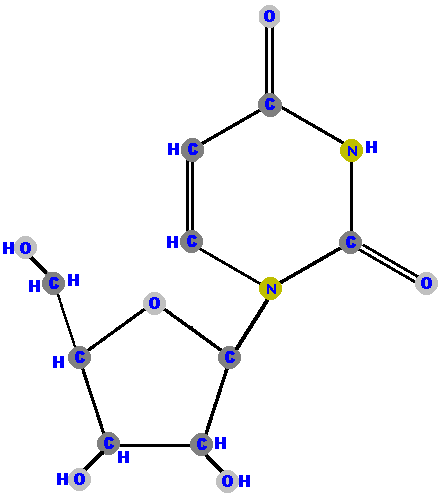 |
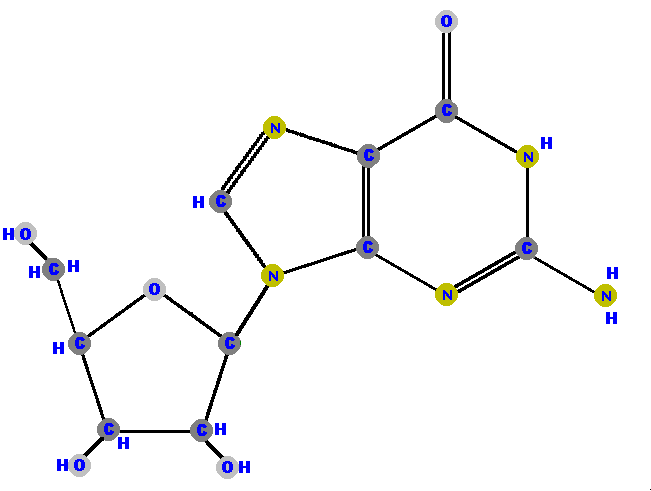 |
 |
Below are the four deoxyribonucleosides. Notice that they do NOT have a hydroxyl group (OH) at the C2' position. Also, notice that thymine is represented among the deoxyribonucleosides instead of uracil.
 |
 |
 |
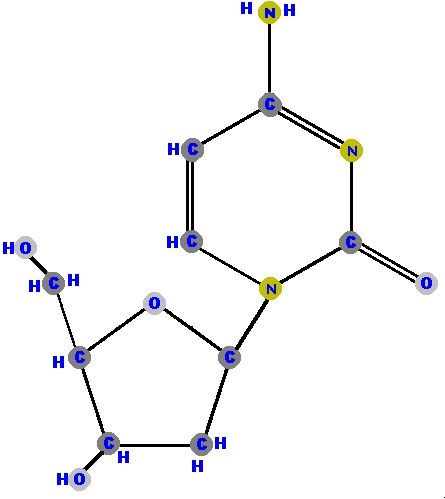 |
Phosphate can be attached to the sugar though the 5' carbon to
give a "triple molecule" of phosphate, sugar and base
called a nucleotide.
[Remember this difference this way. Nucleoside stops
at sugar but nucleotide takes
a phosphate.]
Nucleotides are the fundamental "units" of the larger
molecules of DNA and RNA.
Adenosine 5'-phosphate is adenosine with a phosphate on the 5' carbon - because that's the only place you can put the phosphate on the ribose. Adenosine 5'-phosphate is called adenylate, and the other nucleotides are renamed with that "-ylate" ending too. I will shortly list these names for you to study in a logical manner but for now, here are the four ribonucleotides (made of ribose with a phosphate at the 5' carbon and one of the four possible bases on the 1' carbon).
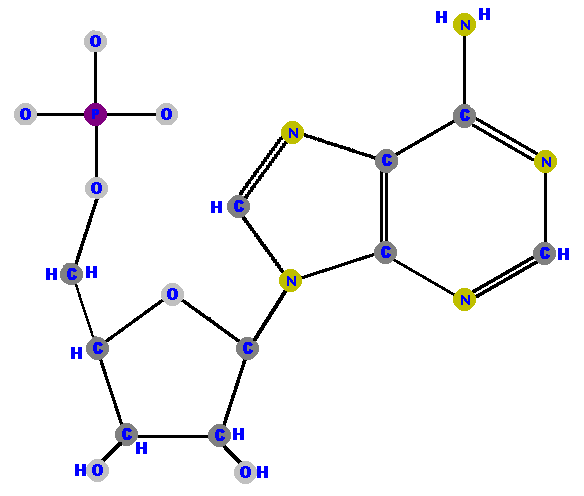 |
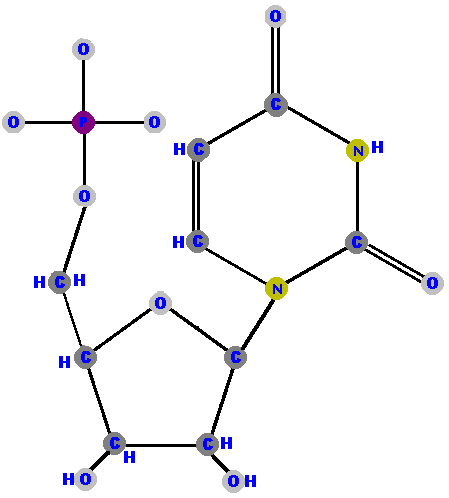 |
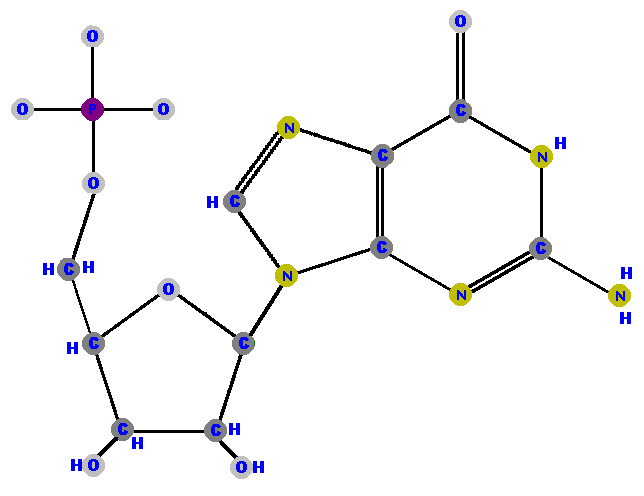 |
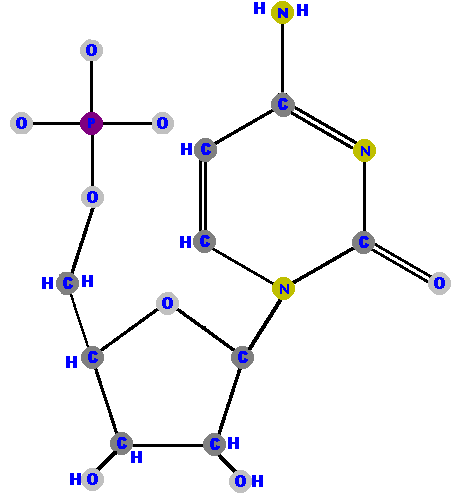 |
Now here are the four deoxyribonucleotides (made of deoxyribose with a phosphate at the 5' carbon and one of the four possible bases on the 1' carbon).
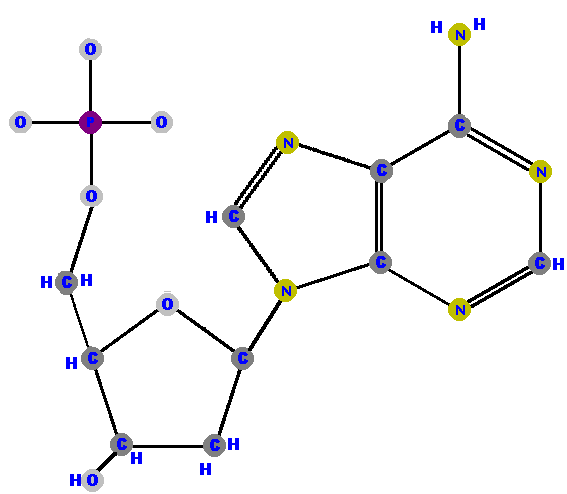 |
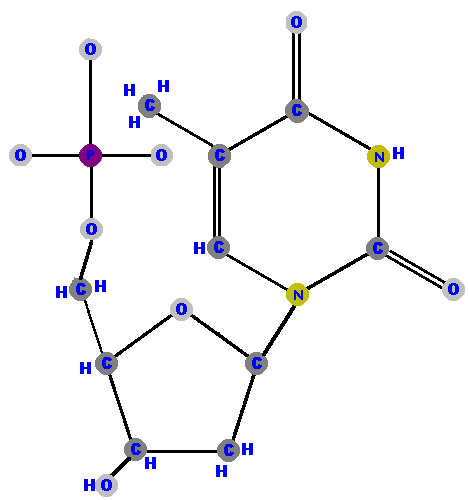 |
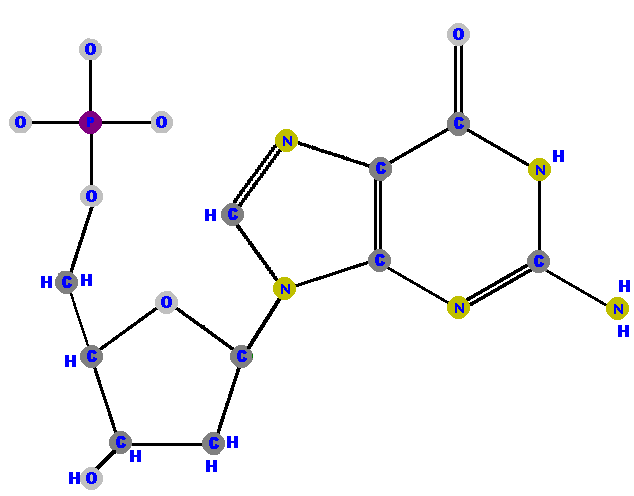 |
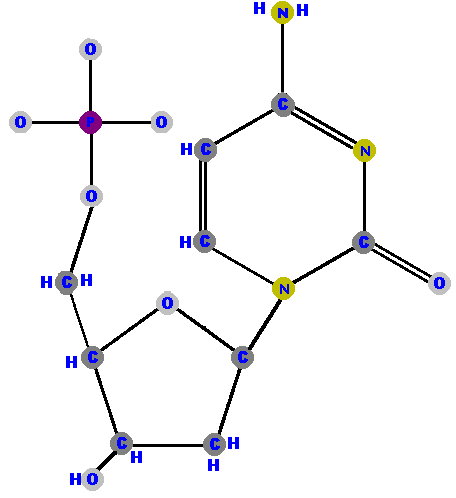 |
It is important to understand that I have been forced to draw
three dimensional molecules on a two dimensional surface. (I am
not sophisticated enough an artist to render molecules as if they
were in three dimensions.) Also, molecules are very "dynamic"
and tend to rotate around bonds unless constrained by multiple
bond or ring structures. For example, the deoxythymidylate molecule above would
tilt and rotate at some points in the third dimension so as
to move the methyl group (CH3) away from the phosphate. Indeed,
I have had to slightly distort the phosphate group in that molecule in order that
it not appear as if it were overlapping the methyl group.
As we now move to the most complicated structures, keep in mind
that some spacing and fine positioning might be slightly distorted
from the reality of the molecules.
Here is a review of all the strange new names you have seen so far. Get a "feel" for them and, once you have the basics in your head, the names of all of them should come very easy. I have also listed the common abbreviations because you will be seeing a lot of them soon. Notice the "deoxys" get a "d" prefix. Carefully read through this table and absorb it.
|
| (5'-monophosphate) | |
(5'-monophosphate) | ||
Before we get to the polymers, linked units of DNA and RNA, I
want to explain that nucleotides are sometimes called "nucleoside
monophosphates" because that is exactly what they are - nucleoside
monophosphates - a nucleoside with a single phosphate.  Don't get
confused here. It is still a nucleoside (because it is the base
and a sugar) BUT it is attached to a phosphate so it should be
called a nucleotide. (Right?) However, it is also correct to call
a nucleotide a "nucleoside phosphate". Why the two different
names for the same molecule? Because sometimes we like to use
the name to describe the molecule in a slightly different way
so as to explain the process it performs or undergoes.
Don't get
confused here. It is still a nucleoside (because it is the base
and a sugar) BUT it is attached to a phosphate so it should be
called a nucleotide. (Right?) However, it is also correct to call
a nucleotide a "nucleoside phosphate". Why the two different
names for the same molecule? Because sometimes we like to use
the name to describe the molecule in a slightly different way
so as to explain the process it performs or undergoes.
Nucleoside diphosphates have a pair of phosphates (PO3-PO4) and
nucleoside triphosphates have a triplet of phosphates (PO3-PO3-PO4)
attached to the 5' carbon of the sugar. All three (nucleoside
monophosphates, nucleoside diphosphates and nucleoside triphosphates)
are nucleotides but, unless we renamed them according to the number
of phosphates, it would not be obvious that these molecules have
different numbers of phosphates. The number of phosphates is important
in certain biological reactions and structures. For example ....
All nucleoside triphosphates are high energy and unstable but once they lose their second and third phosphates (ATP->ADP + P ->AMP + P) the remaining nucleoside monophosphate is very stable and can no longer provide any energy. (I may be getting a head of myself but now is a good time to tell you that it is the energy released when the free floating nucleoside triphosphates become nucleoside monophosphates that allows the DNA or RNA to link into long polymers.)
By now, students who have had a good education in chemistry will
have noticed that I am "cheating" with the phosphate
structures by not showing the double bonds, hydrogens or charges.
That's because I have tried to keep this lesson and drawings simple.
In point of fact, each phosphate has one or two double bonds which
"resonate" to produce a complex "whirlwind"
of electrons. That is too complicated for us, at this level, but
I will tell you that this complex bonding contributes to the high
energy stored in the pyrophosphate linkages. (Don't worry about
it.  ) However, I must remind you that each phosphate
carries one or two negative charges. That's because, in order
to complete their electron complement (to complete their outer
shell, as chem students would say) the phosphates should have
one or two hydrogens bonded to them. Indeed, with those hydrogens
in place, chem students would recognize the phosphates as phosphoric
acid (H3PO4). When placed in water, some of the hydrogens drift
away and take with them their positive charge. That leaves the
phosphate with an "unbalanced" electron complement -
or put more simply, a negative charge.
) However, I must remind you that each phosphate
carries one or two negative charges. That's because, in order
to complete their electron complement (to complete their outer
shell, as chem students would say) the phosphates should have
one or two hydrogens bonded to them. Indeed, with those hydrogens
in place, chem students would recognize the phosphates as phosphoric
acid (H3PO4). When placed in water, some of the hydrogens drift
away and take with them their positive charge. That leaves the
phosphate with an "unbalanced" electron complement -
or put more simply, a negative charge.
Why is this important? Well, it explains why DNA and RNA have the "A". The "A" stands for acid. (DNA is deoxyribonucleic acid. RNA is ribonucleic acid.) A molecule that releases hydrogens is said to be an acid (by definition) and by releasing the hydrogens it takes on a negative charge (by the laws of electrostatics). Note that, until now, I have been talking about bases (on the 1' carbon) so you might have easily though that DNA and RNA were bases! They are not. They are acids. The bases in DNA and RNA are only "weak bases" because they only weakly attract hydrogens. (Molecules that attract hydrogens, even weakly, are called "bases".) If there were no phosphates involved, these molecules would be bases not acids.
Take home message - the phosphates in DNA and RNA cause these molecules to have a net negative charge and is the reason they are acids.
These drawings are very useful for understanding the structure, function and replication of DNA but it would be a waste of time and energy to represent the entire molecule, especially anything much longer (trinucleotides, quadnucleotides, etc). And it would be unnecessary to represent all that detail because the only part of the molecule that changes, along its length, is the sequence of the bases. Indeed, rather than draw the bases it is far more useful to simply list their abbreviations. Biochemists use several different ways to diagram or write the polynucleotides. In order to fit these diagrams across a page, they are represented horizontally - instead of vertically (which is how I've draw them above). So, imagine we rotate those drawings by 90 degrees in order to make them look better.
| One method uses a stick diagram to remind us of the zigzag backbone of the sugar-phosphates. Notice that the terminal OH, represented on the 3' extreme, is the only OH in the backbone and is displayed prominently because biochemists need to know that the OH is there (and available for subsequent polymerization with more nucleotides - more on that later). | 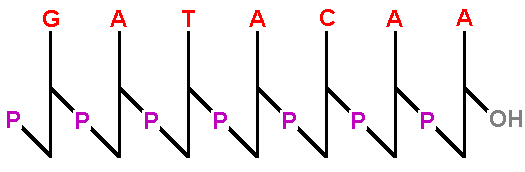 |
| This zigzag diagram is useful but cannot be typed so a simpler representation is to type the sequence with the phosphates and bases abbreviated, all in a straight line. |  |
Notice that this simpler representation shows all the phosphates
including the one at the 5' end. More importantly, the other side of the molecule does NOT have a phosphate so it
is inferred that it must have a hydroxyl (OH) there. That means
we can still figure out the over all direction of the molecule
from its 5' (phosphate, PO4) on the left to its 3' (hydroxyl, OH) end on the right. Of course
the problem with this second representation is that it wastes
a lot of space and time writing in those phosphates when the only
important phosphate is the phosphate at the 5'
end.
However, if we removed all the "p's" we would not
know which end has the inferred hydroxyl end and we could not
orient the molecule so we would not know which end is the 5' end
and which end is the 3' end. (Yes, it is very important to know
the orientation, as you will learn shortly.)
| A third and very abbreviated representation is possible if we accept a rule that, sequences of bases are always written such that the left side is the 5' end (with a dangling phosphate) and the right hand side is the 3' end (with the OH group). |  |
This last representation is simple and easy to write and is the best way to write the sequence of a polynucleotide. The only rule to remember is that ALL sequences are written from the 5' to the 3' direction. (This shorthand is so well accepted that there is even a biotech company called "Five Prime Three Prime" whose logo is simply 5'->3')
You now know how to draw polynucleotides, understand their orientation
and read sequences.  The sequences represent the long strand of
polynucleotides and you can easily "read" the sequence
5' to 3' (from left to right). However, you may be surprised to
learn that DNA is rarely in the form of a single strand of polynucleotides.
It is usually composed of a pair of strands!
The sequences represent the long strand of
polynucleotides and you can easily "read" the sequence
5' to 3' (from left to right). However, you may be surprised to
learn that DNA is rarely in the form of a single strand of polynucleotides.
It is usually composed of a pair of strands!
A fellow named Chargaff discovered that the DNA from any particular cell (or overall, in any tissue or organism), has equal amounts of purines and pyrimidines. That is, there is a one-to-one correspondence between purines (A and G) and pyrimidines (T and C). Indeed (specifically), the amount of adenine (A) equals that amount of thymine (T) and the amount of guanine (G) equals the amount of cytosine (C). This rule is so important and so well documented - in all living things, from bacteria to man - that it has been called "Chargaff's rule".
In 1953, using structural information (called "X-ray diffraction" and created by Avery five years earlier) and with the help of Rosalind Parkers to explain that structural information, Watson and Crick proposed a structure for DNA that simultaneously explained why Chargaff's rule "rules", how DNA can replicate and also hinted at how it might encode the genetic information!
The remainder of this lesson will teach you about the structure of genomic DNA.
Study this diagram because there are a lot of useful details in
it. (Click on the image to get the big picture then print a copy so you can study it.) I have had to twist and spin some bases in order to show the positioning (just as the molecules do) and I have left the lettering also rotated so you can appreciate the complexity introduced (and
to help orient you).
I have drawn blue dotted lines to represent the hydrogen bonds. Let's discuss them in detail in order to help you understand this important concept.
Note that each nucleotide is attached to the next nucleotide by a phosphate group (PO4) linking the 5' carbon (5'C) of one sugar to the 3' carbon (3'C) of the next sugar. That should be no surprise because we just talked about that.
|
Now take a good look at this simplier diagram and notice that the strand
on the right is in an opposite orientation to the one on the left.
That is - the stand on the right has a 3' at the top and a 5'
at the bottom but the strand on the left is the other way around
(with a 5' at the top and a 3' at the bottom). Don't get confused
here! All polynucleotides are written in the 5' to 3' direction.
Think back to the "sugar man" drawing and superimpose it on this more complicated structure. You will see (imagine) that the chain of sugar men on the left are positioned standing "up right" but the ones on the right are "upside down", standing on their heads! We say the stands are "antiparallel" meaning they run in opposite directions. All double-stranded DNA is antiparallel because that is the only way to match the bonding made by the bases. | 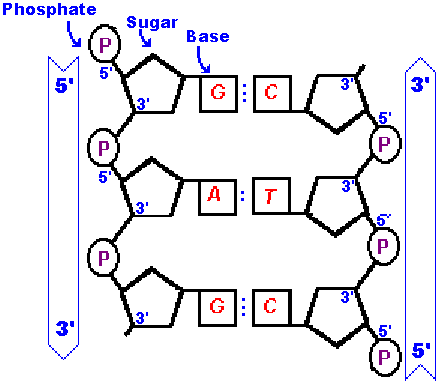 |
Some things are so important that they should be repeated - so here goes. Besides, it allows me to (re)introduce some ideas that you may have missed.
|
Confused? Think about the sugar men as you take in this simplified
diagram.
Imagine the sugar men on the left are all standing on the shoulders of each other. Each has a base (left hand) connected to the C1 (left shoulder). In order for the other strand of sugar man to "shake left hands" they must flip over on their heads to line up correctly. (Well, it is not exactly like shaking hands because you and I don't have to stand that way to shake hands but the sugar men do not have the flexibility that you and I have in our shoulders. Think about it.) Therefore, the two strands are antiparallel and the bases "complement" each other, via hydrogen bonds and specific shapes, giving rise to Chargaff's rule. |  |
The phosphodiester bonds and sugars also have a restricted orientation
that cause them to twist a small amount in order to line up with
the opposite, complementary strand. The result is that the two
strands wrap around each other in a double helix. 
|
Notice that either strand, imagined on its own, rotates as it
moves up or down like a helical staircase.
]People commonly call such staircases "spiral staircases" but they are wrong. A spiral changes its radius as you move along its length but helixes maintain the same radius. Only fancy mansions can afford truly spiral staircases because they take up so much room! In the case of DNA, a spiral structure would be a disaster because the bases would not be able to pair with each other.] Also notice that each strand will maintain its 5' to 3' direction and be antiparallel to its opposite strand but, due to the helix, it is hard to track the direction. Before, without the helix, it was easy to see that the strand on the left ran from 5' to 3' as you went from top to bottom and the strand on the right was in the opposite orientation - making it easy to understand. However, the helix causes the strands to switch from left to right - but they still maintain their original 5' to 3' orientation because the molecules do not change. As you can see, there are a variety of ways to illustrate this complicated molecule and we can pick and choose among various diagrams depending upon what level of complexity we are trying to imagine. You need complete atom-by-atom models when first learning the structures in order to appreciate the higher level complications, such as why stands run 5' to 3' and antiparallel, but at other levels simple drawings suffice. Indeed, the sugar phosphate "backbone" is so boring that it is often represented as a simple ribbon and, if need be, labeled 5' to 3'. | 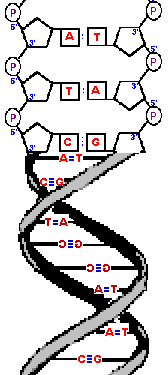 |
|
At the other extreme - a more realistic model, called a "space
filling model", shows the size of each atom and, as you can
see, makes the whole things very difficult to interpret. (Don't
worry about the color-coding used.)
With thanks to Eric Martz for his permission to use this image. Please visit his website at http://molvis.sdsc.edu/dna. These space filling models give the most accurate representation of the DNA molecule - but, of course the real molecule is much smaller. How small?
Well, that depends on the length of the DNA.
| 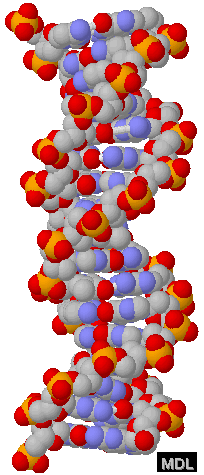 |
|
While we are on the subject, it is worth mentioning that adjacent
bases are separated by 0.34 nanometers along the helix axis and
rotated 36 degrees from each other.
(What I mean by that is the helix turns such that, if you were looking down the "tube" you would see that the bases are 36 degrees away from each other. I think there are more than enough drawings in this lesson so I won't illustrate that.)
Due to that rotation, the helical structure
repeats after ten nucleotides.
An extremely advanced student might note that DNA forms a "right-handed helix", meaning the twist goes from the lower left to the upper right as you move along its length. This is an "advanced" property of this complicated molecule but it is not uncommon for some textbooks and even good scientists to accidentally switch the image (by making a "mirror" image). Yes- it's a complicated molecule! However, you (too) can understand this "handedness" property. | 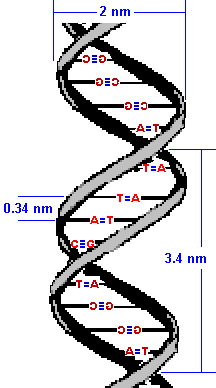 |
|
Here are three images of DNA.
The one on the left is the molecule you have seen before and correctly oriented so the A, T, G and C are correct (but as the molecule twists around you see the letter from behind so they look reversed). That is normal right-handed DNA. Notice that if you run your finger along any strand on your side of the molecule (lighter colored) that the path ALWAYS goes from lower left to upper right. That is the property that makes this a right-handed molecule. The second image is the same molecule rotated upside down. (See how all the letters are upside down?) Notice that this is still a right-handed helix! That is because the "handedness" of a molecule does not depend upon its positioning - it is a property of the molecule itself. Now look at the third image. I've used a little trick (called "reflection") to make a mirror image of the original molecule. This molecule on the right is a left-handed helix and is not the normal, natural form of DNA. Notice how its twists the wrong way. Run your finger along all three of these molecules in order to appreciate the handedness of DNA. | (Correct) 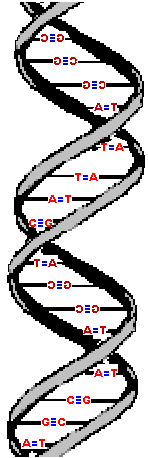 | (Upside down but OK) 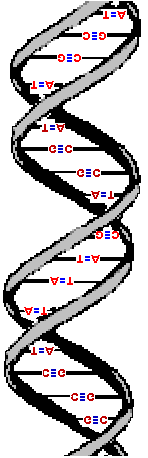 | (WRONG!) 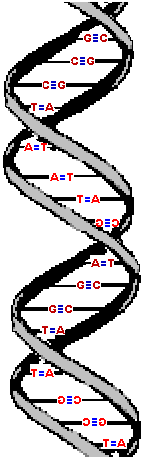 |
You've learned enough detail for today! We've covered a lot of very important information in this lesson and it is different from the kind of things we discussed in previous courses. However, it is important that you completely understand this material in order to get the most out of the rest of this course. Study it well and work through the SAQs to solidify your understanding. Once you are comfortable with "what DNA is" I will tell you "what DNA does" in our next lesson! | 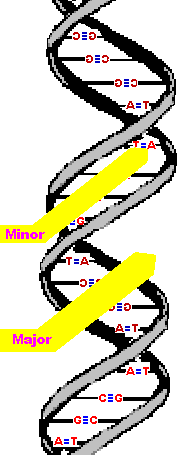
|
This work was created by Dr Jamie Love  and
and  licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
licensed under a Creative Commons Attribution-ShareAlike 4.0 International License.
| Table of Contents | Homepage | How to get a FREE copy of the entire course (hypertextbook) | Frequently Asked Questions |